This week in MathOnco 248
Clinical translation of adaptive therapy, stackelberg evolutionary game theory, adaptive PARP inhibitor treatment, mutation fixation, somatic evolution, and more.
“This week in Mathematical Oncology” — Mar. 30, 2023
> mathematical-oncology.org
From the editor:
Today we feature articles on clinical translation of adaptive therapy, stackelberg evolutionary game theory, adaptive PARP inhibitor treatment, mutation fixation, somatic evolution, and more.
Enjoy,
Jeffrey West
jeffrey.west@moffitt.org
“I have deeply regretted that I did not proceed far enough at least to understand something of the great leading principles of mathematics; for men thus endowed seem to have an extra sense."
- Darwin
A survey of open questions in adaptive therapy: Bridging mathematics and clinical translation
Jeffrey West, Fred Adler, Jill Gallaher, Maximilian Strobl, Renee Brady-Nicholls, Joel Brown, Mark Roberson-Tessi, Eunjung Kim, Robert Noble, Yannick Viossat, David Basanta, Alexander RA AndersonStackelberg evolutionary game theory: how to manage evolving systems
Alexander Stein, Monica Salvioli, Hasti Garjani, Johan Dubbeldam, Yannick Viossat, Joel S. Brown and Kateřina StaňkováNeuroblastoma arises in early fetal development and its evolutionary duration predicts outcome
Verena Körber, Sabine A. Stainczyk, Roma Kurilov, Kai-Oliver Henrich, Barbara Hero, Benedikt Brors, Frank Westermann & Thomas HöferGeneralizable biomarker prediction from cancer pathology slides with self-supervised deep learning: A retrospective multi-centric study
Jan Moritz Niehues, Philip Quirke, Nicholas P. West, Alexander Brobeil, Michael Hoffmeister, Jakob Nikolas Kather
Adaptive therapy for ovarian cancer: An integrated approach to PARP inhibitor scheduling
Maximilian Strobl, Alexandra L. Martin, Jeffrey West, Jill Gallaher, Mark Robertson-Tessi, Robert Gatenby, Robert Wenham, Philip Maini, Mehdi Damaghi, Alexander AndersonMutant fixation in the presence of a natural enemy
Dominik Wodarz, Natalia L. KomarovaLearning the functional landscape of microbial communities
Abigail Skwara, Karna Gowda, Mahmoud Yousef, Juan Diaz-Colunga, Arjun S Raman, Alvaro Sanchez, Mikhail Tikhonov, Seppe KuehnClonal differences underlie variable responses to sequential and prolonged treatment
Dylan L Schaff, Aria J Fasse, Phoebe E White, Robert J Vander Velde, Sydney M ShafferSomatic Evolution of Cancer: A New Synthesis
Ulfat Baing, Rohini Kharate, Milind WatveLong-range interactions and disorder facilitate pattern formation in spatial complex systems
Fabrizio Olmeda, Steffen Rulands
12th Annual Southern California Systems Biology Conference
#SoCalSysBio23: The conference will provide an opportunity for diverse research groups across Southern California institutions to showcase their current research activities in all areas of Systems Biology. The meeting format will consist of short faculty talks, with lightning talks and poster sessions for trainees. Come share your latest work, build community, and stimulate collaborations! Registration deadline: April 15 (click here to register).
Deconvolution of clinical variance in CAR-T pharmacology and response
Daniel Kirouac: “Combining mathematical modelling, bioinformatics, and machine learning to understand the pharmacology of T cell therapeutics”
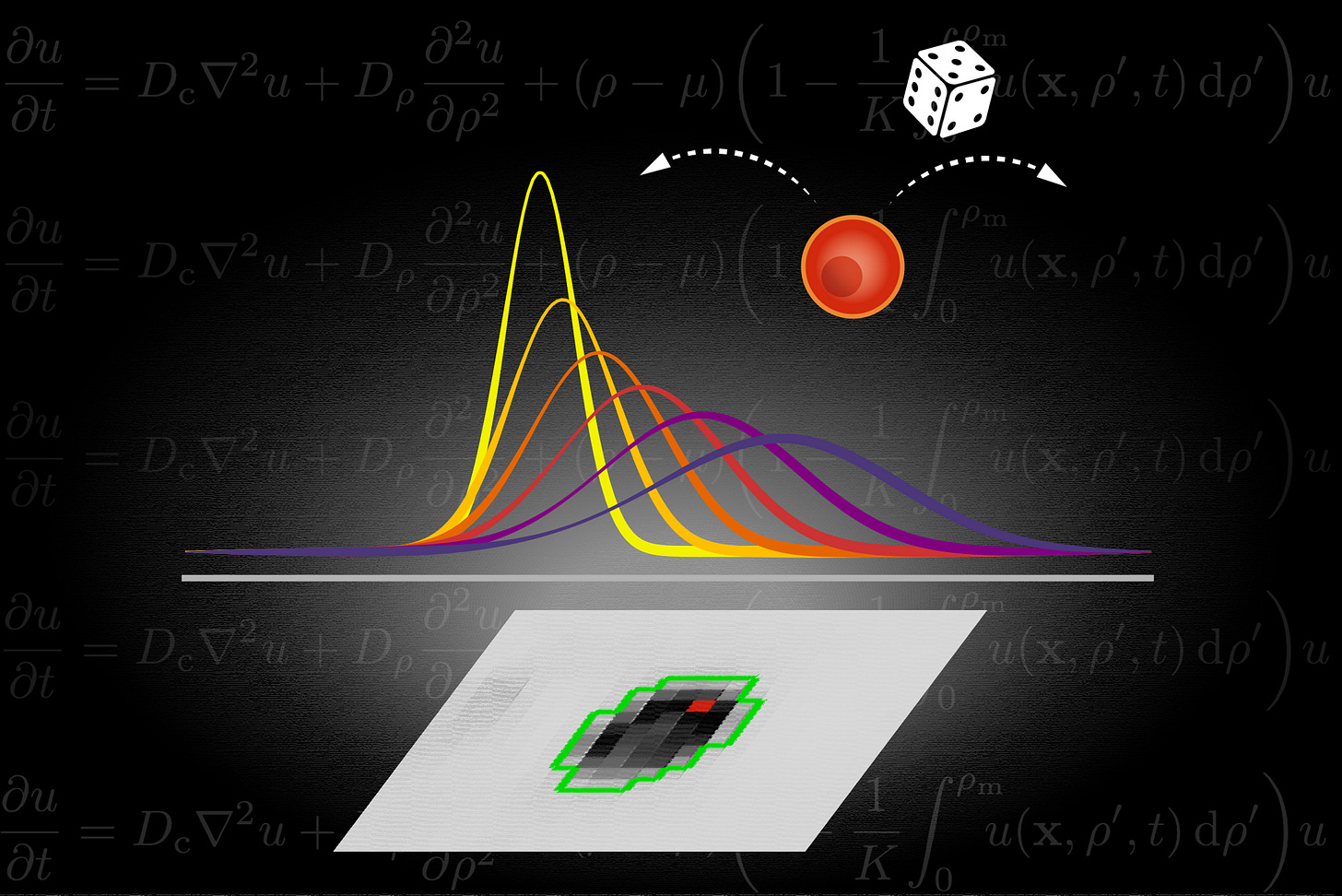
The newsletter now has a dedicated homepage where we post the cover artwork for each issue. We encourage submissions that coincide with the release of a recent paper from your group. This week’s artwork:
Based on the paper: Metabolic activity grows in human cancers pushed by phenotypic variability in iScience.
Artist: Jesús J. Bosque (@JesusJBosque)
Caption: “PET scans provide information of paramount importance to the clinician, but are also a powerful tool to access tumour metabolism for the researcher. Using pre-treatment PET images from patients with cancer, we identified how the hotspot of metabolic activity (marked in red in the image) reflects underlying processes that drive the tumour to more aggressive states. To model it, we used a PDE model phenotypically structured on the proliferation, where cells can randomly change their proliferation rate upwards or downwards with equal probability. The model shows that this variability progressively shifts the proliferation profiles to higher values, what is depicted in the image by the curves showing the successive distributions. Interestingly, the model described very well the traits identified in the data, therefore, we discuss that random phenotypic changes suffice to fuel the observed increases in tumour metabolic activity.”
Visit the mathematical oncology page to view jobs, meetings, and special issues. We will post new additions here, but the full list can found at mathematical-oncology.org.
1. Jobs
Current subscriber count: >1.6k