This week in MathOnco 257
Deterministic evolution, transcriptional hallmarks, growth-inhibition, reaction-diffusion, and more.
“This week in Mathematical Oncology” — June 8, 2023
> mathematical-oncology.org
From the editor:
Today we feature articles on deterministic evolution, transcriptional hallmarks, growth-inhibition, reaction-diffusion, and more.
Enjoy,
Jeffrey West
jeffrey.west@moffitt.org
Deterministic evolution and stringent selection during preneoplasia
Kasper Karlsson, Moritz J. Przybilla, Eran Kotler, Aziz Khan, …, Carlos J. Suarez, Chris P. Barnes, Calvin J. Kuo & Christina CurtisHallmarks of transcriptional intratumour heterogeneity across a thousand tumours
Avishai Gavish, Michael Tyler, Alissa C. Greenwald, Rouven Hoefflin, …, Amit Tirosh, Mario L. Suvà, Sidharth V. Puram & Itay TiroshEarly Decision Making in a Randomized Phase II Trial of Atezolizumab in Biliary Tract Cancer Using a Tumor Growth Inhibition-Survival Modeling Framework
Colby S. Shemesh, Phyllis Chan, Mathilde Marchand, Antonio Gonçalves, Shweta Vadhavkar, Benjamin Wu, Chunze Li, Jin Y. Jin, Stephen P. Hack, Rene BrunoApproximating the Value of Zero-Sum Differential Games with Linear Payoffs and Dynamics
Jeroen Kuipers, Gijs Schoenmakers & Kateřina StaňkováModeling Tumour Growth with a Modulated Game of Life Cellular Automaton Under Global Coupling
Vladimir García-Morales, José A. Manzanares & Javier Cervera
The reaction-diffusion basis of animated patterns in eukaryotic flagella
James Cass, Hermes Bloomfield-GadêlhaA Comparison of Mutation and Amplification-Driven Resistance Mechanisms and Their Impacts on Tumor Recurrence
Aaron Li, Danika Kibby, Jasmine FooAn Agent-Based Model of Monocyte Differentiation into Tumour-Associated Macrophages in Chronic Lymphocytic Leukemia
Nina Verstraete, Malvina Marku, Marcin Domagala, Hélène Arduin, Julie Bordenave, Jean-Jacques Fournié, Loïc Ysebaert, Mary Poupot, Vera PancaldiNon-genetic differences underlie variability in proliferation among esophageal epithelial clones
Raul A Reyes Hueros, Rodrigo A Gier, Sydney M Shaffer
Point of View: Beware ‘persuasive communication devices’ when writing and reading scientific articles
eLife: Olivier Corneille Is a corresponding author, Jo Havemann, Emma L Henderson, Hans IJzerman, Ian Hussey, Jean-Jacques Orban de Xivry, Lee Jussim, Nicholas P Holmes, Artur Pilacinski, Brice Beffara, Harriet Carroll, Nicholas Otieno Outa, Peter Lush, Leon D Lotter
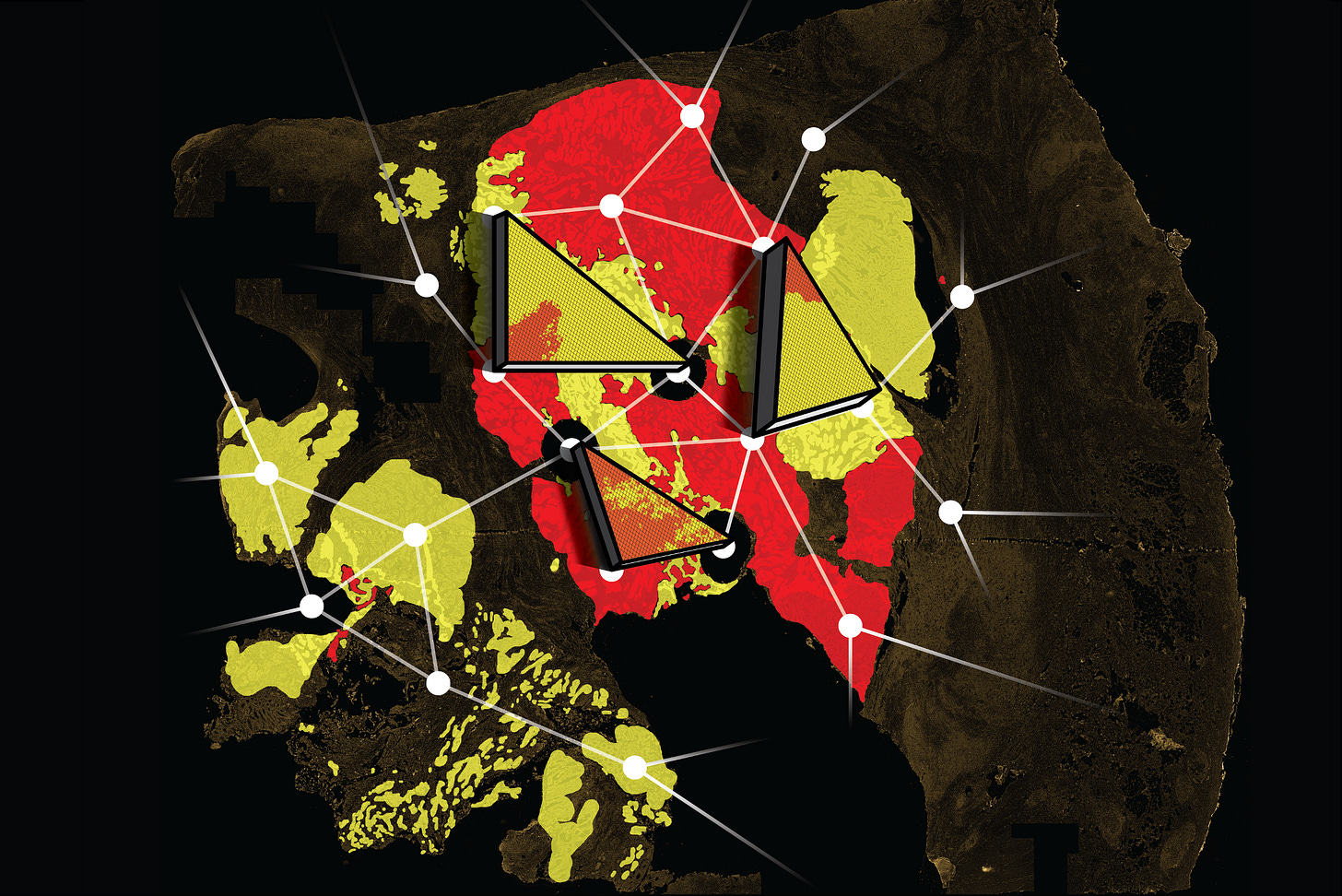
The newsletter now has a dedicated homepage where we post the cover artwork for each issue. We encourage submissions that coincide with the release of a recent paper from your group. This week’s artwork:
Based on the paper: First passage time analysis of spatial mutation patterns reveals sub-clonal evolutionary dynamics in colorectal cancer in PLoS Computational Biology
Artist: Magnus Haughey, Ann-Marie Baker & Weini Huang
Caption: "How do geometrical patterns of tumour sub-populations relate to their underlying dynamics? In our recent study, we set out to tackle the problem of how to quantify tumour sub-clonal dynamics in single time-point human colorectal cancer (CRC) samples using only the information contained within the spatial architecture of different tumour sub-populations. By simulating random walks on the pixels of high-resolution images of human CRC, we exploited the first passage times between the wild-type and mutant KRAS, BRAF, and PIK3CA populations to quantify their observed geometry. Comparing these to similar measurements of simulated spatial tumours, generated using a spatial agent-based model, we then estimated the parameters of our model which best recapitulated the observed sub-population architectures in patient samples. Our results suggested that, despite containing almost no genetic information, it is still possible to extract some information about tumour sub-clonal dynamics by analysing the shape of cell sub-populations."
Visit the mathematical oncology page to view jobs, meetings, and special issues. We will post new additions here, but the full list can found at mathematical-oncology.org.
1. Jobs
Current subscriber count: >1.6k